Two years ago, I bought a Garmin Forerunner 305 to records pulse and location during my training sessions. pytrainer does a good job graphing sporting excursions such as running or cycling sessions, but with a little bit of R code, you can do your own analysis and graphing. Most of the code needed is already written and compiled into nice high-level libraries for R, eg the plotKML, sp, zoo, and XML packages.
Instructions for installing plotKML,
There are two ways to get the data from the device, each has its own package
gpsbabel is convenient to use, but it does not extract the total distance which garmin fore runner 305 records at each point. While latitude and longitude plus elevation can be used to derive this distance, garmin-forerunner-tools provides this data directly from the device. The only drawback of garmin-forerunner-tools is that the procedure needed to get data from the device into R has an extra step.
Instructions for installing gpsbabel
system("sudo gpsbabel -t -i garmin -f usb: -o gpx,garminextensions -F ~/my.gpx") library(XML) my.xml.tree <- xmlParse(file="my.gpx")
The alternative is using the commands in the package garmin-forerunner-tools.
my.dir <- system("mktemp -d", intern = TRUE) setwd(my.dir) system("sudo garmin_save_runs") system("sudo chown -R hans:hans .") my.files <- list.files(pattern=".gmn", recursive=T) my.xml.data <- system(paste("garmin_dump", my.files[6]), intern = TRUE) ## Reading only the session in file number 6. my.xml.tree <- htmlTreeParse(my.xml.data) r <- xmlRoot(my.xml.tree)[[1]] ## xmlattrs(xmlchildren(r)[[2]])
my.activity <- xmlAttrs(xmlChildren(r)[[1]])[['sport']] my.session <- xmlAttrs(xmlChildren(r)[[1]])[['track']] my.start.time <- as.POSIXct(xmlAttrs(xmlChildren(r)[[2]])[['start']], format = "%Y-%m-%dT%H:%M:%S") ## duration is in form HH:MM:SS.S my.duration <- xmlAttrs(xmlChildren(r)[[2]])[['duration']] ## distance is in meters xmlAttrs(xmlChildren(r)[[2]])[['distance']] ## waypoint data are in nodes named "point" these <- which(names(sapply(xmlChildren(r), names)) == "point") ## throw away the last waypoint, since it misses most of the variables these <- these[-length(these)] my.mat <- matrix(NA, ncol = length(xmlAttrs(xmlChildren(r)[[these[1]]])), nrow = length(these)) ##for(i in 1:550){ for(i in these[-length(these)]){ my.mat[i-(these[1]-1),] <- xmlAttrs(xmlChildren(r)[[i]]) } my.df <- data.frame(time.stamp = as.POSIXct(my.mat[,2], format = "%Y-%m-%dT%H:%M:%S"), lat = as.numeric(my.mat[,3]), lon = as.numeric(my.mat[,4]), altitude = as.numeric(my.mat[,5]), distance = as.numeric(my.mat[,6]), heart.rate = as.integer(my.mat[,7])) my.df$dt.time <- c(0, my.df$time.stamp[-1] - my.df$time.stamp[-length(my.df$time.stamp)]) my.df$dt.distance <- c(0, my.df$distance[-1] - my.df$distance[-length(my.df$distance)]) my.df$speed <- c(0, my.df$dt.distance[-1]/my.df$dt.time[-1]) * 3.6 ## Absolute time library(zoo) my.ts <- zoo(my.df$heart.rate, my.df$time.stamp) plot(my.ts, xlab = "time", ylab = "pulse", main = as.Date(rownames(as.data.frame(my.ts[1])))) grid() ## Relative time, custom time labels my.steps <- c(0, 3600 * as.numeric(tail(my.df$time.stamp, 1) - head(my.df$time.stamp, 1)) / 4 / 60 * 1:4) my.hour <- floor(my.steps / 60) my.minute <- floor(my.steps - 60 * floor(my.steps / 60)) my.lab <- paste(my.hour, formatC(my.minute, flag = "0", width = 2), sep = ":") plot(y = my.df$heart.rate, my.df$time.stamp - my.df$time.stamp[1], xaxt = "n", ylab = "pulse", xlab = "time", type = "l") axis(side = 1, at = c(0, 3600 * as.numeric(tail(my.df$time.stamp, 1) - head(my.df$time.stamp, 1)) / 4 * 1:4), labels = my.lab) grid()
B* Animation
xpathSApply(my.xml.tree,'//ns:name', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1")) [1] "11" "12" "13" "14" "15" "16" "17" "18" "19" "20" "21" "22" "23" "24" "25" "26" "27" "28" "29" "30" "31" "32
Let's say we wanted to do a time series analysis of heart rate data from the session named "24". For the time serie analysis, we use the package zoo
library(zoo) my.ts <- zoo(as.numeric(xpathSApply(my.xml.tree, '//*/ns:name[text()[contains(.,"24")]]//following-sibling::ns:trkseg/*/ns:extensions', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1"))), as.POSIXct(strptime(xpathSApply(my.xml.tree, '//*/ns:name[text()[contains(.,"24")]]//following-sibling::ns:trkseg/ns:trkpt/ns:time', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1")), tz = "GMT", format = "%Y-%m-%dT%H:%M:%SZ")))
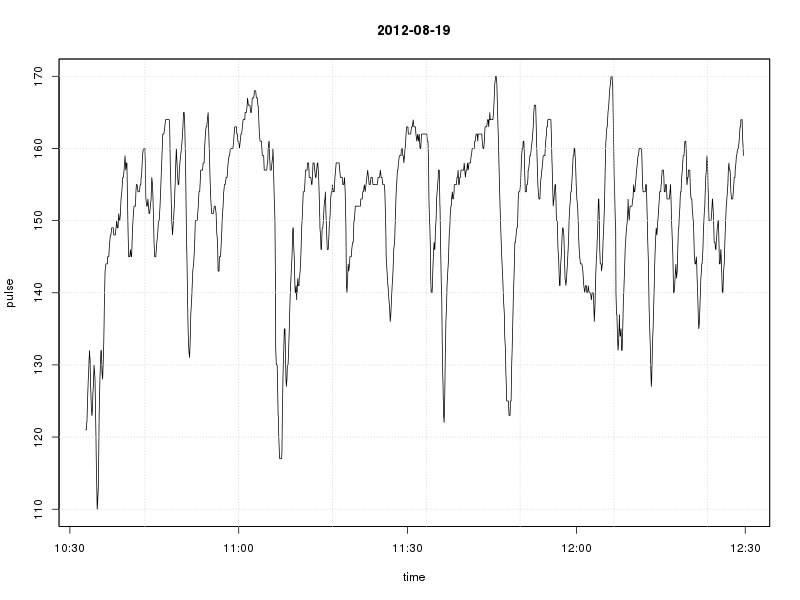
To plot the heart rates over time in this session, we can now simply call plot()
plot(my.ts, xlab = "time", ylab = "pulse", main = as.Date(rownames(as.data.frame(my.ts[1])))) grid()

In order to compare two sessions, the time data in each session must be relative to the starting point of each session - currently the time data are absolute. By substracting the time of the first observation from all observation, we get relative time data.
Eg. Compare session 32 with session 33
my.ts.1 <- zoo(1E11 * as.numeric(xpathSApply(my.xml.tree, '//*/ns:name[text()="40"]//following-sibling::ns:trkseg/*/ns:extensions', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1"))), as.POSIXct(strptime(xpathSApply(my.xml.tree, '//*/ns:name[text()="40"]//following-sibling::ns:trkseg/ns:trkpt/ns:time', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1")), tz = "GMT", format = "%Y-%m-%dT%H:%M:%SZ"))) ## Save the date my.title <- as.Date(index(my.ts.1)[1]) ## use time differences, not time stamps index(my.ts.1) <- index(my.ts.1) - index(my.ts.1)[1] my.ts.2 <- zoo(1E11 * as.numeric(xpathSApply(my.xml.tree, '//*/ns:name[text()="41"]//following-sibling::ns:trkseg/*/ns:extensions', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1"))), as.POSIXct(strptime(xpathSApply(my.xml.tree, '//*/ns:name[text()="41"]//following-sibling::ns:trkseg/ns:trkpt/ns:time', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1")), tz = "GMT", format = "%Y-%m-%dT%H:%M:%SZ"))) index(my.ts.2) <- index(my.ts.2) - index(my.ts.2)[1] my.x.max <- max(c(index(my.ts.1), index(my.ts.2))) my.y.max <- max(my.ts.1, my.ts.2) plot(my.ts.1, xlab = "time", ylab = "pulse", main = my.title, xlim = c(0, my.x.max), ylim = c(50, my.y.max)) lines(my.ts.2, lty = 2)
library(plotKML) library(spacetime) my.df <- data.frame(time = time.stamps <- as.POSIXct(strptime(xpathSApply(my.xml.tree, '//*/ns:name[text()[contains(.,"24")]]//following-sibling::ns:trkseg/ns:trkpt/ns:time', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1")), tz = "GMT", format = "%Y-%m-%dT%H:%M:%SZ"))) pos <- t(xpathSApply(my.xml.tree, '//*/ns:name[text()[contains(.,"24")]]//following-sibling::ns:trkseg/ns:trkpt', xmlAttrs, namespaces = c(ns = "http://www.topografix.com/GPX/1/1"))) pos.df <- data.frame(lon = as.numeric(pos[ ,2]), lat = as.numeric(pos[ ,1])) coordinates(my.df) <- pos.df proj4string(my.df) <- CRS("+proj=longlat +datum=WGS84") plotKML(my.df, folder.name="/tmp/bar", open.kml = FALSE) Plotting the first variable on the list KML file opened for writing... Writing to KML... Closing /tmp/bar.kml Object written to: /tmp/bar.kml
Start google-earth, open /tmp/bar.kml, save as image, and you will get something like this:
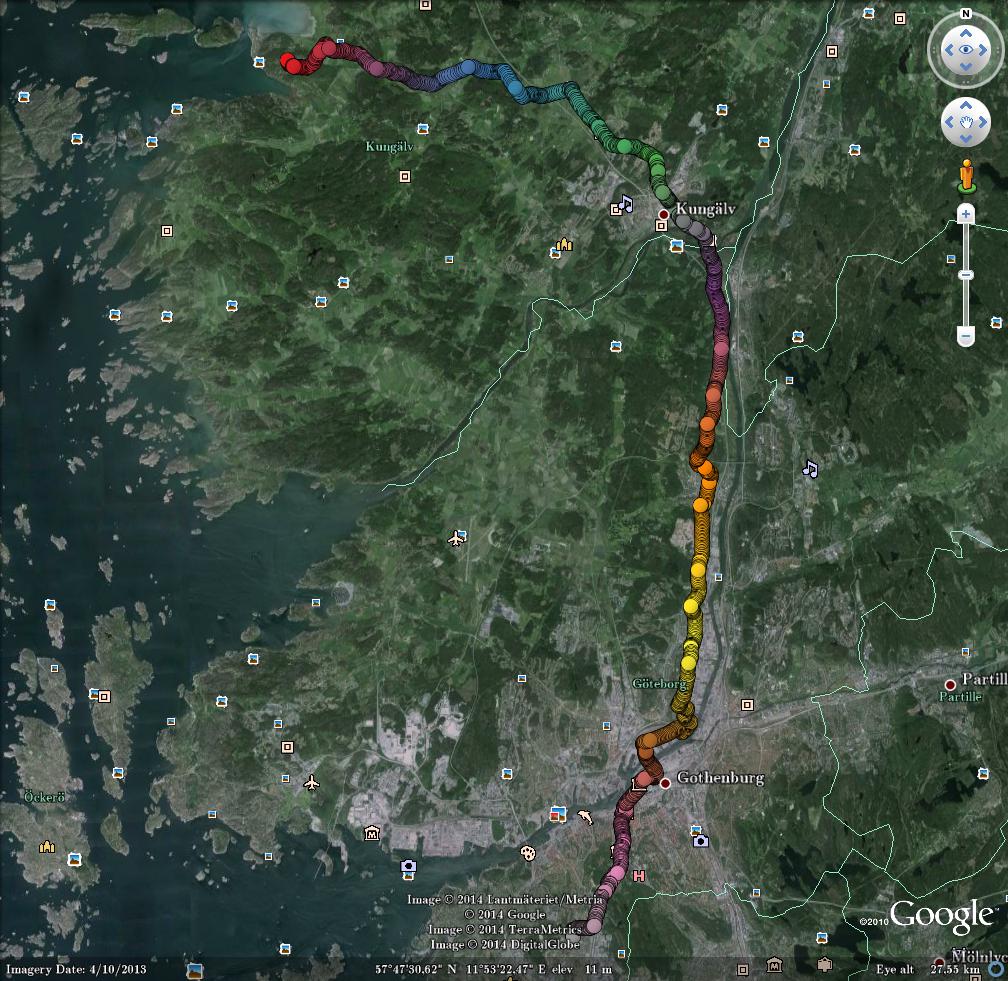
Or just share the kml-file for others to open in google-earth.
library(spacetime) my.gpx <- readGPX("~/my.gpx") ## merge all training sessions into a common data.frame ## This fails unless there are exactly the same numbers of columns in all sessions. my.df <- data.frame() for(i in 1:length(my.gpx$tracks)){ for(j in 1:length(my.gpx$tracks[[i]])){ if(length(my.gpx$tracks[[i]][[j]]) > 0){ my.gpx$tracks[[i]][[j]]$session <- i print(paste(ncol(my.gpx$tracks[[i]][[j]]), "i:", i, "j:", j)) my.df <- rbind(my.df, my.gpx$tracks[[i]][[j]]) } } }
plotKML on Debian Wheezyapt-get install libgdal-dev tk-dev
install.packages("plotKML", "fossil")
To save disk space you may now uninstall the newly installed development libraries. But the runtime library libgdal1 should be kept.
apt-get install libgdal1 apt-get --purge remove libgdal-dev tk-dev apt-get --purge autoremove
sudo apt-get install googleearth-package make-google-earth-package sudo dpkg -i googleearth_6.0.3.2197+0.7.0-1_i386.deb sudo apt-get -f install sudo dpkg -i googleearth_6.0.3.2197+0.7.0-1_i386.deb
apt-get install gpsbabel
apt-get install garmin-forerunner-tools
my.ts.1 <- my.ts <- zoo(as.numeric(xpathSApply(my.xml.tree, '//*/ns:name[text()="33"]//following-sibling::ns:trkseg/*/ns:extensions', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1"))),
as.POSIXct(strptime(xpathSApply(my.xml.tree, '//*/ns:name[text()="33"]//following-sibling::ns:trkseg/ns:trkpt/ns:time', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1")), tz = "GMT", format = "%Y-%m-%dT%H:%M:%SZ"))) Save the date session.1.date <- as.Date(index(my.ts.1)1) use time differences, not time stamps index(my.ts.1) <- index(my.ts.1) - index(my.ts.1)1 my.ts.2 <- my.ts <- zoo(as.numeric(xpathSApply(my.xml.tree, '//*/ns:name[text()="34"]//following-sibling::ns:trkseg/*/ns:extensions', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1"))), as.POSIXct(strptime(xpathSApply(my.xml.tree, '//*/ns:name[text()="34"]//following-sibling::ns:trkseg/ns:trkpt/ns:time', xmlValue, namespaces = c(ns = "http://www.topografix.com/GPX/1/1")), tz = "GMT", format = "%Y-%m-%dT%H:%M:%SZ"))) session.2.date <- as.Date(index(my.ts.2)1) index(my.ts.2) <- index(my.ts.2) - index(my.ts.2)1 my.x.max <- max(c(index(my.ts.1), index(my.ts.2))) my.y.max <- max(my.ts.1, my.ts.2) plot(my.ts.1, xlab = "time (in seconds)", ylab = "pulse", main = "Comparison of training sessions", xlim = c(0, my.x.max), ylim = c(50, my.y.max)) lines(my.ts.2, lty = 2) legend(x = my.x.max / 2.5, y = 60 + (my.y.max - 60) / 2, legend = c(session.1.date, session.2.date), lty = c(1, 2), text.width = strwidth(session.1.date))